Super-resolution microscopy, in light, is a term that gathers several techniques, which allow images to be taken with a higher resolution than the one imposed by the. Due to the, the in conventional light microscopy is limited, as stated (for the special case of widefield illumination) by in 1873. In this context, a diffraction-limited microscope with N.A. And light with wavelength λ reaches a lateral resolution of d = λ/(2 N.A.) - a similar formalism can be followed for the axial resolution (along the optical axis, z-resolution, depth resolution). The resolution for a standard optical microscope in the visible light spectrum is about 200 nm laterally and 600 nm axially. Experimentally, the attained resolution can be measured from the (FWHM) of the (PSF) using images of point-like objects. Although the resolving power of a microscope is not well defined, it is generally considered that a super-resolution microscopy technique offers a resolution better than the one stipulated by Abbe.Super-resolution imaging techniques rely on the (photon tunneling microscopy as well as those that utilize the and ) or on the.
- 3d Deconvolution
- Deconvolution Software For Mac
- Comparison Of Deconvolution Software In Microscopy Science
Among the latter are techniques that improve the resolution only modestly (up to about a factor of two) beyond the diffraction-limit like the (with closed pinhole), or confocal microscopy aided with computational methods such as deconvolution or detector-based pixel reassignment (e.g. Re-scan microscopy, pixel reassignment ), the, and also structured illumination microscopy technologies like SIM and.There are two major groups of methods for super-resolution microscopy in the far-field that can improve the resolution with a much larger factor:. Deterministic super-resolution: The most commonly used emitters in biological microscopy, show a nonlinear response to excitation, and this nonlinear response can be exploited to enhance resolution. These methods include, and SSIM. Stochastic super-resolution: The chemical complexity of many molecular light sources gives them a complex temporal behavior, which can be used to make several close-by fluorophores emit light at separate times and thereby become resolvable in time.
These methods include (SOFI) and all single-molecule localization methods (SMLM) such as, FPALM, STORM and dSTORM.On October 8, 2014, the was awarded to, and for 'the development of super-resolved,' which brings ' into the '. Contents.History In 1978, the first theoretical ideas had been developed to break the using a as a confocal laser scanning fluorescence microscope where the light is focused ideally from all sides to a common focus that is used to scan the object by 'point-by-point' excitation combined with 'point-by-point' detection.Some of the following information was gathered (with permission) from a chemistry blog's review of sub-diffraction microscopy techniques. For a review, see also reference.In 1986, the super-resolution optical microscope based on stimulated emission was patented by Okhonin. Super-resolution techniques Photon tunneling microscopy (PTM).
Comparison of the resolution obtained by (top) and 3D structured illumination microscopy (3D-SIM-Microscopy, bottom). Shown are details of a. Nuclear pores (anti-NPC) red, nuclear envelope (anti-) green, (-staining) blue. Scale bar: 1µm.There is also the wide-field (SIM) approach.SI enhances spatial resolution by collecting information from frequency space outside the observable region. This process is done in reciprocal space: The (FT) of an SI image contains superimposed additional information from different areas of reciprocal space; with several frames with the illumination shifted by some, it is possible to computationally separate and reconstruct the FT image, which has much more resolution information. The reverse FT returns the reconstructed image to a super-resolution image.A number of applications have recently surfaced, in which SIM microscopy could potentially replace as part of routine diagnosis.
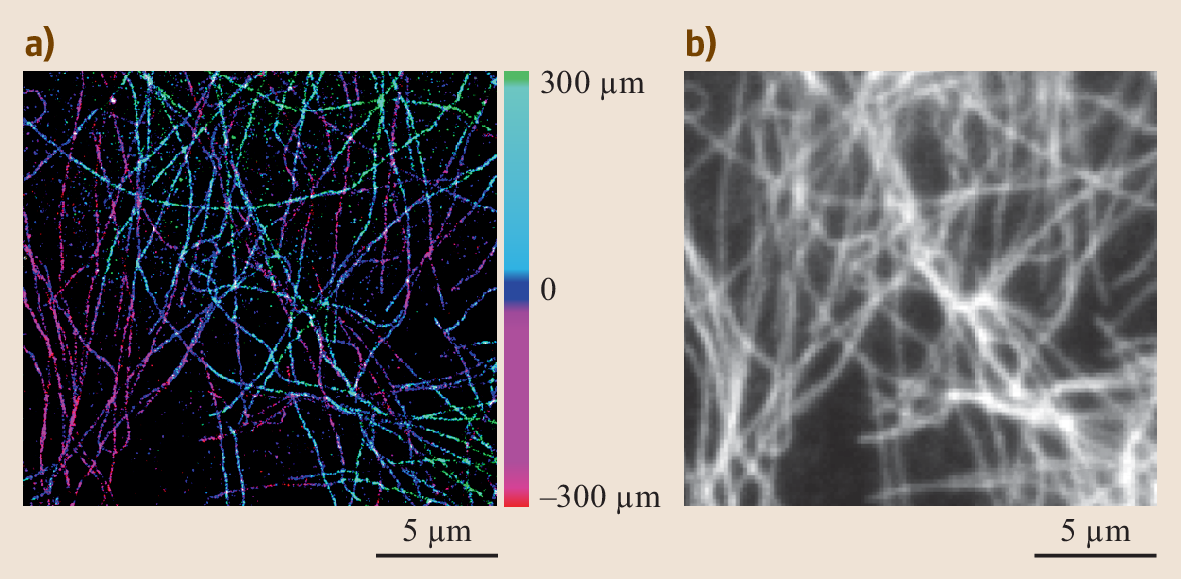
These include diagnosis of kidney disorders, kidney cancer, and blood diseases.Although the term 'structured illumination microscopy' was coined by others in later years, Guerra (1995) first published results in which light patterned by a 50 nm pitch grating illuminated a second grating of pitch 50 nm, with the gratings rotated with respect to each other by an angular amount to achieve magnification. Although the illuminating wavelength was 650 nm, the 50 nm grating was easily resolved. This showed a nearly 5X improvement over the Abbe resolution limit of 232 nm that should have been the smallest obtained for the N.A. And wavelength used. In an extension and further development of this work, Guerra showed that super-resolved lateral topography is attained by phase-shifting the evanescent field. Patents were issued to Guerra individually or with colleagues and assigned to Polaroid Corp. Licenses to this technology were procured by Dyer Energy Systems, Calimetrics Inc.
And then Nanoptek Corp. For use of this super-resolution technique in optical data storage and microscopy.
Images of and stages recorded with 3D-SIM. SMI + of human eye tissue affected byOne implementation of structured illumination is known as spatially modulated illumination. Like standard structured illumination, the SMI technique modifies the (PSF) of a microscope in a suitable manner.
In this case however, 'the optical resolution itself is not enhanced'; instead structured illumination is used to maximize the precision of measurements of fluorescent objects, to 'enable size measurements at molecular dimensions of a few tens of nanometers'.The microscope achieves structured illumination by using one or two opposing interfering laser beams along the axis. The object being imaged is then moved in high-precision steps through the wave field, or the wave field itself is moved relative to the object by phase shift. This results in an improved axial size and distance resolution.SMI can be combined with other super resolution technologies, for instance with 3D LIMON or LSI- as a interferometer with laterally structured illumination (this last instrument and technique is essentially a phase-shifted photon tunneling microscope, which employs a total internal reflection light microscope with phase-shifted evanescent field (Guerra, 1996) ).
This SMI technique allows one to acquire light-optical images of autofluorophore distributions in the sections from human eye tissue with previously unmatched optical resolution. Use of three different excitation wavelengths (488, 568 and 647 nm), enables one to gather spectral information about the autofluorescence signal. This has been used for human eye tissue affected. Deterministic functional techniques microscopy is an with very high resolution that can image details in samples that cannot be imaged with conventional. Within RESOLFT the principle of and are generalized.
Also, there are techniques with other concepts than RESOLFT or SSIM, for example using optical property of. Stimulated emission depletion (STED). Single YFP molecule super resolution microscopy / SPDMphymodA single, tiny source of light can be located much better than the resolution of a microscope: Although the light will produce a blurry spot, computer algorithms can be used to accurately calculate the center of the blurry spot, taking into account the of the microscope, the noise properties of the detector, and so on. However, this approach does not work when there are too many sources close to each other: The sources all blur together.SPDM (spectral precision distance microscopy) is a family of techniques in which gets around this problem by measuring just a few sources at a time, so that each source is 'optically isolated' from the others (i.e., separated by more than the microscope's resolution, typically 200-250 nm). Then, the above technique (finding the center of each blurry spot) can be used.If the molecules have a variety of different spectra (absorption spectra and/or emission spectra), then it is possible to look at light from just a few molecules at a time by using the appropriate light sources and filters.
3d Deconvolution
Molecules can also be distinguished in more subtle ways based on and other techniques.The structural resolution achievable using SPDM can be expressed in terms of the smallest measurable distance between two in their spatial position determined punctiform particle of different spectral characteristics ('topological resolution'). Modeling has shown that under suitable conditions regarding the precision of localization, particle density etc., the 'topological resolution' corresponds to a ' which in terms of the classical definition is equivalent to a much improved optical resolution.SPDM is a localization microscopy which achieves an effective optical resolution several times better than the conventional optical resolution (approx.
200-250 nm), represented by the half-width of the main maximum of the effective point image function. By applying suitable laser optical precision processes, position and distances significantly smaller than the half-width of the point spread function (conventionally 200-250 nm) can be measured with nanometer accuracy between targets with different spectral signatures. An important area of application is genome research (study of the functional organization of the ). Another important area of use is research into the structure of membranes.SPDMphymod. Dual color localization microscopy SPDMphymod/super-resolution microscopy with GFP & RFP fusion proteinsLocalization microscopy for many standard fluorescent dyes like, and molecules is possible if certain photo-physical conditions are present.
With this so-called (physically modifiable fluorophores) technology a single laser wavelength of suitable intensity is sufficient for nanoimaging in contrast to other localization microscopy technologies that need two laser wavelengths when special photo-switchable/photo-activatable fluorescence molecules are used. A further example for the use of SPDMphymod is an analysis of (TMV) particles or the study of.Based on singlet triplet state transitions it is crucial for that this process is ongoing and leading to the effect that a single molecule comes first into a very long-living reversible dark state (with half-life of several seconds even) from which it returns to a fluorescent state emitting many photons for several milliseconds before it returns into a very long-living so-called irreversible dark state. SPDMphymod microscopy uses fluorescent molecules that are emitting the same spectral light frequency but with different spectral signatures based on the flashing characteristics. By combining two thousands images of the same cell, it is possible using laser optical precision measurements to record localization images with significantly improved optical resolution.Standard fluorescent dyes already successfully used with the technology are GFP, RFP, YFP, Alexa 488, Alexa 568, Alexa 647, Cy2, Cy3, Atto 488 and fluorescein.Cryogenic Optical Localization in 3D (COLD). Cryogenic Optical Localization in Three Dimensions (COLD) allows to determine the four biotin binding sites in the protein streptavidin.Cryogenic Optical Localization in 3D (COLD) is a method that allows localizing multiple fluorescent sites within a single small- to medium-sized biomolecule with Angstrom-scale resolution. The localization precision in this approach is enhanced because the slower photochemistry at low temperatures leads to higher number of photons that can be emitted from each fluorophore before photobleaching.
As a result, cryogenic stochastic localization microscopy reaches the required sub-molecular resolution to resolve the 3D positions of several fluorophores attached to a small protein. By employing algorithms known from electron microscopy, the 2D projections of fluorophores are reconstructed into a 3D configuration. COLD brings fluorescence microscopy to its fundamental limit given by the size of the label. The method can also be combined with other structural biology techniques such as X-ray crystallography, magnetic resonance spectroscopy and electron microscopy to provide valuable complementary information and specificity.BALM.
FBALM Super-resolution single molecule localisation microscopy using DNA structure fluctuation assisted binding activated localisation microscopyBALM (Binding Activated Localization Microscopy) is a general concept for Single Molecule localization Microscopy (SMLM) super-resolved imaging of DNA-binding dyes based on modifying the properties of DNA and the dye. By careful adjustment of the chemical environment leading to local, reversible DNA melting and hybridization control over the fluorescence signal of the DNA-binding dye molecules can be introduced. Intercalating and minor-groove binding DNA dyes can be used to register (optically isolate) only a few DNA-binding dye signals at a time. DNA structure fluctuation-assisted BALM (fBALM) has been used to nanoscale differences in nuclear architecture with an anticipated structural resolution of approximately 50 nm.
Imaging chromatin nanostructure with binding-activated localization microscopy based on DNA structure fluctuations. Recently, significant enhancement of fluorescence quantum yield of NIAD-4 upon binding to was exploited to BALM imaging of amyloid fibrils and oligomers. STORM, PALM and FPALM.
Main article:Stochastic optical reconstruction microscopy (STORM), photo activated localization microscopy (PALM) and fluorescence photo-activation localization microscopy (FPALM) are super-resolution imaging techniques that utilize sequential activation and time-resolved localization of photoswitchable fluorophores to create high resolution images. During imaging, only an optically resolvable subset of fluorophores is activated to a fluorescent state at any given moment, such that the position of each fluorophore can be determined with high precision by finding the centroid position of the single-molecule images of particular fluorophore. The fluorophore is subsequently deactivated, and another subset is activated and imaged. Iteration of this process allows numerous fluorophores to be localized and a super-resolution image to be constructed from the image data. These three methods were published independently during a short period of time and their principle is identical. STORM was originally described using Cy5 and Cy3 dyes attached to nucleic acids or proteins, while PALM and FPALM was described using photoswitchable fluorescent proteins.
In principle any photoswitchable fluorophore can be used, and STORM has been demonstrated with a variety of different probes and labeling strategies. Using stochastic photoswitching of single fluorophores, such as Cy5, STORM can be performed with a single red laser excitation source. The red laser both switches the Cy5 fluorophore to a dark state by formation of an adduct and subsequently returns the molecule to the fluorescent state. Many other dyes have been also used for STORM. In addition to single fluorophores, dye-pairs consisting of an activator fluorophore (such as Alexa 405, Cy2, and Cy3) and a photoswitchable reporter dye (such as Cy5, Alexa 647, Cy5.5, and Cy7) can be used for STORM. In this scheme, the activator fluorophore, when excited near its absorption maximum, serves to reactivate the photoswitchable dye to the fluorescent state.
Multicolor imaging has been performed by using different activation wavelengths to distinguish dye-pairs based on the activator fluorophore used or using spectrally distinct photoswitchable fluorophores either with or without activator fluorophores. Photoswitchable fluorescent proteins can be used as well. Highly specific labeling of biological structures with photoswitchable probes has been achieved with antibody staining, direct conjugation of proteins, and genetic encoding.STORM has also been extended to three-dimensional imaging using optical astigmatism, in which the elliptical shape of the point spread function encodes the x, y, and z positions for samples up to several micrometers thick, and has been demonstrated in living cells.
Deconvolution Software For Mac
To date, the spatial resolution achieved by this technique is 20 nm in the lateral dimensions and 50 nm in the axial dimension and the temporal resolution is as fast as 0.1–0.33s. Points accumulation for imaging in nanoscale topography (PAINT) Points accumulation for imaging in nanoscale topography (PAINT) is a single-molecule localization method that achieves the stochastic single-molecule fluorescence by molecular adsorption/absorption and photobleaching/desorption. The first dye used is which is nonfluorescent in aqueous solution but is fluorescent when inserted into a hydrophobic environment such as micelles or living cell walls. Thus when the concentration of the dye is small, in the order of nanomolar level, such that the molecule's rate to the diffraction-limited area is in the millisecond region. The stochastic binding of single dye molecules (probes) to an immobilized target can be spatially and temporally resolved under a typical widefield fluorescence microscope.

Each dye is photobleached to return the field to dark for the next dye to bind and be observed. The advantage of this method comparing to other stochastic method is in addition to obtaining the super-resolved image of the fixed target, it can measure the dynamic binding kinetics of the diffusing probe molecules in the solution to the target.
Combining with 3D super-resolution technique (e.g. The double-helix point spread function develop in Moerner's group) and using photo-activated dyes, power-dependent active intermittency and points accumulation for imaging in nanoscale topography (SPRAIPAINT) can super-resolve live-cell walls. PAINT works by maintaining a balance between the dye adsorption/absorption and photobleaching/desorption rates. This balance can be estimated with stochastic statistical principles. The or rate of a dilute solute to a surface or interface in a (gas or liquid) solution can be calculated using. The photobleaching/desorption rate can be measured for a given solution condition and illumination power density.PAINT has been further extended to regular dyes termed as DNA-PAINT when the dynamic binding and unbinding of a dye-labeled DNA probe to a fixed is used to achieve the stochastic single-molecule imaging. DNA-PAINT is no longer limited to the environment-sensitive dyes and can measure both the adsorption and the desorption kinetics of the probes to the target.
The method utilizes the camera blurring effect of moving dyes. When a regular dye is diffusing in the solution, its image on a typical CCD camera is blurred because of its relatively fast moving speed and the relatively long camera exposure time, thus it contributes to the fluorescence background. However, when it binds to a fixed target, the dye stop moving and a clear point spread function can be taken by the camera. Thus, a general term for this modification is mbPAINT. When a (TIRF) is used to imaging, the excitation depth is limited to 100 nm from the substrate, which further reduces the fluorescence background from the blurred dyes near the substrate and the background in the bulk solution. Very bright dyes can be used for mbPAINT which gives typical single-frame spatial resolutions 20 nm and single-molecule kinetic temporal resolution 20 ms under relatively mild photoexcitation intensities, which is useful in studying molecular separation of single proteins.The temporal resolution has been further improved (20 times) using a rotational phase mask placed in the Fourier plane during data acquisition and resolve the distorted point spread function that contains temporal information.
The method was named Super Temporal-Resolved Microscopy (STReM). Label-free localization microscopy. Main article:It is possible to circumvent the need for PSF fitting inherent in single molecule localization microscopy (SMLM) by directly computing the temporal autocorrelation of pixels. This technique is called super-resolution optical fluctuation imaging (SOFI) and has been shown to be more precise than SMLM when the density of concurrently active fluorophores is very high.Omnipresent Localization Microscopy (OLM) Omnipresent Localisation Microscopy (OLM) is an extension of Single Molecule Microscopy (SMLM) techniques that allow high-density single molecule imaging with an incoherent light source (such as Mercury arc lamp) and a conventional epifluorescence microscope setup.
A short burst of deep blue excitation (350-380 nm instead of 405 nm laser) can for a prolonged reactivation of molecules. A resolution of 90 nm on test specimens.
Finally, correlative STED and SMLM imaging can be performed on the same biological sample using a simple imaging medium, which can provide a basis for a further enhanced resolution. These findings can democratise superresolution imaging and help any scientist to generate high-density single molecule images even with a limited budget.Combination of techniques 3D light microscopical nanosizing (LIMON) microscopy. 3D Dual Colour Super Resolution Microscopy with Her2 and Her3 in breast cells, standard dyes: Alexa 488, Alexa 568 LIMON3 D LIMON (Light MicrOscopical nanosizing microscopy) images using the microscope are made possible by the combination of and, whereby first the SMI and then the SPDM process is applied.The process determines the center of particles and their spread in the direction of the microscope axis. While the center of particles/molecules can be determined with a 1–2 nm precision, the spread around this point can be determined down to an axial diameter of approx. 30–40 nm.Subsequently, the position of the individual particle/molecule is determined using SPDM, achieving a precision of a few nanometers.As a biological application in the 3D dual color mode the spatial arrangements of Her2/neu and Her3 clusters was achieved. The positions in all three directions of the protein clusters could be determined with an accuracy of about 25 nm.
Integrated correlative light and electron microscopy Combining a super-resolution microscope with an electron microscope enables the visualization of contextual information with the labelling provided by fluorescence markers. This overcomes the problem of the black backdrop that the researcher is left with when using only a light microscope. In an integrated system, the sample is measured by both microscopes simultaneously.
Comparison Of Deconvolution Software In Microscopy Science
Enhancing of techniques using neural networks Recently, owing to advancements in artificial intelligence computing, deep learning neural networks have been used for super-resolution enhancing of photographic images, optical microscopy from 40x to 100x, 20x optical microscope to 1500x scanning electron microscope via a neural lens, positron-emission tomography and fluorescence microscopy. See also. (MUM). (STED). (PALM). (STORM).References.